Finally, VHHs have enhanced stability and solubility and are easily expressed and purified from E. coli or yeast expression systems, which may make production cheaper and more efficient. We previously isolated three VHHs from a naive llama phagedisplay VHH library against a�CCbtx, the most potent a neurotoxin from the venom of N. kaouthia. Since their affinities were too low for therapeutic use, we set out, in this study, to isolate higher affinity single-domains from a hyperimmune library. We immunized a llama with crude N. kaouthia venom instead of purified a�CCbtx. Our immunization scheme was inspired by a “low dose, low volume, multi-site immunization” protocol Vorinostat described by Chotwiwatthanakun et al. that showed potent horse IgG titres for antivenom production. However, we modified this protocol 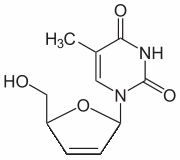 in terms of adjuvant used, time interval between injections, total number of injection sites, and injection volumes. Our llama, which was immunized with up to 2 mg of crude N. kaouthia, did not suffer from any visible signs cobra envenomation or local reaction at the injection sites. After receiving the first boost, the llama generated a rapid humoral response to N. kaouthia venom and more specifically to a�CCbtx. Others have also observed a rapid rise in IgG titres following venom-immunzation in camelids and horses. Thereafter, we observed an abrupt decrease in the humoral response after the second boost. In subsequent injections, we decided to replace the adjuvant with incomplete Freund��s adjuvant, and consequently observed the humoral response to increase again. One report has shown that a�CCbtx elicits its highest titres rapidly in the immunization schedule and that after reaching a plateau, titres slowly decrease even after continued injections. Our llama’s final antiserum titre to N. kaouthia was 3.06105 and is very comparable to that observed by Cook et al. in camels immunized with N. nigricollis venom. After detecting a llama HCAb immune response against a�CCbtx, we constructed a large phage-displayed VHH library of 5.06109 clones with 84% containing a VHH coding sequence. Sequence analysis revealed that the 3rd round of panning generated several unique VHH sequences. Based on CDR homology and CDR3 length, the VHH clones were grouped into either Cluster I or Cluster II. Binders clustered together share high CDR sequence PF-4217903 c-Met inhibitor identity; however, little CDR homology is shared between these two clusters. Furthermore, the a�CCbtx binders from Cluster I have a CDR3 length of 17 amino acid residues while those from Cluster II have 14 residues. Several differences were also noted when these VHHs were aligned against those we previously isolated from a na? ��ve library. The na? ��ve binders all belong to VHH subfamily 1 while the immune ones belong to VHH subfamily 2. As expected, the immune VHHs share little CDR homology with the na? ��ve ones, since they had the opportunity to go through natural affinity maturation. For instance, immune VHHs have a shorter CDR3 region than the na? ��ve ones. This result is consistent with the literature since VHH subfamily 2 have on average a shorter CDR3 region than other subfamilies. We chose two clones from Cluster I and two clones from Cluster II for further characterization. The clones were expressed in 1 L E. coli cultures and purified from the periplasmic fractions using IMAC. Kinetic analysis by SPR revealed that high affinity a�CCbtx binders were isolated from the immune library. Accordingly, Vu et al. have previously suggested that, in contrast to human and mice VHs, amino acid residues from the CDR1 of camelid VHHs play a critical role in antigen-binding. Furthermore, our immune a�C Cbtx VHH binders had,1000-fold higher affinity compared to those we previously isolated from a naive VHH library. Although immune libraries are cumbersome and timeconsuming to construct.
in terms of adjuvant used, time interval between injections, total number of injection sites, and injection volumes. Our llama, which was immunized with up to 2 mg of crude N. kaouthia, did not suffer from any visible signs cobra envenomation or local reaction at the injection sites. After receiving the first boost, the llama generated a rapid humoral response to N. kaouthia venom and more specifically to a�CCbtx. Others have also observed a rapid rise in IgG titres following venom-immunzation in camelids and horses. Thereafter, we observed an abrupt decrease in the humoral response after the second boost. In subsequent injections, we decided to replace the adjuvant with incomplete Freund��s adjuvant, and consequently observed the humoral response to increase again. One report has shown that a�CCbtx elicits its highest titres rapidly in the immunization schedule and that after reaching a plateau, titres slowly decrease even after continued injections. Our llama’s final antiserum titre to N. kaouthia was 3.06105 and is very comparable to that observed by Cook et al. in camels immunized with N. nigricollis venom. After detecting a llama HCAb immune response against a�CCbtx, we constructed a large phage-displayed VHH library of 5.06109 clones with 84% containing a VHH coding sequence. Sequence analysis revealed that the 3rd round of panning generated several unique VHH sequences. Based on CDR homology and CDR3 length, the VHH clones were grouped into either Cluster I or Cluster II. Binders clustered together share high CDR sequence PF-4217903 c-Met inhibitor identity; however, little CDR homology is shared between these two clusters. Furthermore, the a�CCbtx binders from Cluster I have a CDR3 length of 17 amino acid residues while those from Cluster II have 14 residues. Several differences were also noted when these VHHs were aligned against those we previously isolated from a na? ��ve library. The na? ��ve binders all belong to VHH subfamily 1 while the immune ones belong to VHH subfamily 2. As expected, the immune VHHs share little CDR homology with the na? ��ve ones, since they had the opportunity to go through natural affinity maturation. For instance, immune VHHs have a shorter CDR3 region than the na? ��ve ones. This result is consistent with the literature since VHH subfamily 2 have on average a shorter CDR3 region than other subfamilies. We chose two clones from Cluster I and two clones from Cluster II for further characterization. The clones were expressed in 1 L E. coli cultures and purified from the periplasmic fractions using IMAC. Kinetic analysis by SPR revealed that high affinity a�CCbtx binders were isolated from the immune library. Accordingly, Vu et al. have previously suggested that, in contrast to human and mice VHs, amino acid residues from the CDR1 of camelid VHHs play a critical role in antigen-binding. Furthermore, our immune a�C Cbtx VHH binders had,1000-fold higher affinity compared to those we previously isolated from a naive VHH library. Although immune libraries are cumbersome and timeconsuming to construct.
It was necessary to construct an immune library to isolate high affinity binders against
Leave a reply